Ammonia-oxidizing bacteria (AOB) often fail to nitrify at the bulk pH value of the acid soil from which they were isolated. Therefore, ureolysis has long been proposed to explain nitrification in acidic soil in which ammonia concentration is too low to support bacterial ammonia oxidation. Recent discovery has suggested that ammonia-oxidizing archaea (AOA) in culture is able to grow on urea as an energy source (Tourna et al. 2011. PNAS 108: 8420). In addition, the acidophilic Nitrososphaera viennensis was obtained in an acidic soil suggesting that AOA play a key role in nitrifying acidic soil (Lehtovirta-Morley et al., 2011. PNAS. 108: 15892). However, it remains unclear whether ureolysis could afford greater advantage for archaeal ammonia oxidation in acid soil.
Dr. Zhongjun Jia’s group from the Institute of Soil Science, Chinese Academy of Sciences has found that nitrification of archaeal ammonia oxidizers in acidic soil is supported by hydrolysis of urea. (1) By using 15N-isotope tracing technique, LU Lu and coworkers demonstrated that nitrification is supported by hydrolysis of urea at a concentration of 5 µg N per gram acidic soil. This suggests that ammonia oxidation could occur through urea hydrolysis in tea orchard soil (pH 3.75) and forested soil (pH 5.40) under in situ conditions. (2) The high-throughput pyrosequencing at the whole microbial community level provides an almost unbiased strategy to rapidly screen ammonia oxidizers associated with nitrification activity changes in complex soils. It indicates that the relative proportion of archaeal population increased up to 32-fold upon the hydrolysis of urea in acidic soil. (3) Sequencing analysis indicates that there was no bacterial ammonia oxidation and urea-linked nitrification is most likely related to AOA affiliated with an obligate acidophile Nitrosotalea devanaterra.
These findings have been published online by The ISME Journal (Nitrification of archaeal ammonia oxidizers in acid soils is supported by hydrolysis of urea. http://dx.doi.org/10.1038/ismej.2012.45). It has implication for the reassessment of nitrogen cycling in acidic soil that comprises 30% of the world's soils, including more than 50% of potential arable land.
 |
 |
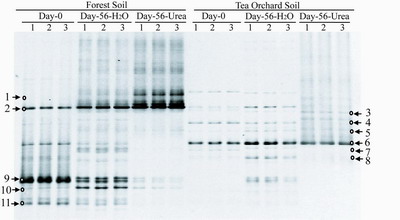
| DGGE fingerprints of archaeal amoA genes in soils. 1, 2, and 3 represent the three samples from triplicate microcosms for each treatment. Day-56-H2O and Day-56-Urea indicate that the soil microcosms received sterilized water or urea once a week, respectively. The arrows indicate the DGGE bands excised for sequencing |
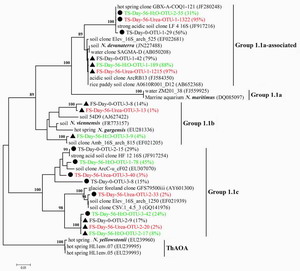
Phylogenetic tree showing the relationship ofhigh-throughput archaeal 16S rRNAgene sequence reads in two acidic soils to those obtained from the GenBank. FS and TS indicate forest soil in red and tea orchard soil in blue with symbols ▲ and ●, respectively. FS-Day-56-Urea-OTU-1-185 (56%) indicates that OTU-1 of archaeal 16S rRNA genes contains 185 sequence reads with >97% identity and accounts for 56% of the total archaeal 16S rRNA reads in forest soil after incubation with urea for 56 days. One representative sequence from each OTU was extracted using the mothur software package for tree construction. Bootstrap values higher than 70% are indicated at branch nodes |
